CFDE Catalyzes Discovery by Breaking Down Data Silos
The lack of data integration limited the transformative potential of the NIH Common Fund (CF) programs. Addressing this opportunity, the NIH Common Fund Data Ecosystem (CFDE) aims to catalyze discovery by breaking the data silos and establishing infrastructure, tools, and practices that enable research combining data, information, and computable knowledge generated by individual programs. CFDE activities included high levels of interactions between participating groups and thematically focused working groups. Collectively, these efforts are improving the Findability, Accessibility, Interoperability, and Reusability (or FAIRness) of CF data and resources. To learn more, view a short overview of the Common Fund Data Ecosystem.
Partnership projects pursued by the CFDE Data Coordinating Centers (DCCs) and the CFDE Coordination Center (CFDE CC) have prepared a series of videos to demonstrate the progress in CFDE. These videos highlight the integrative infrastructure, cross-cutting tools, and new pathways to discovery developed during the pilot phase.
A. Infrastructure and Tools for Data Integration across CFDE Projects
Portal Overview
A portal for navigation of cross-project data using a flexible metadata standard for describing experimental resources in biomedicine and related fields, known as the Crosscut Metadata Model (C2M2).
Data Submission Tools and Processes
A common tool for data submission.
C2M2 Compatibility Test
A tool that allows submitters to the CFDE portal to test C2M2 compatibility of data packages.
Content Registry and Knowledge Base
A tool for coherent registration of resources Content Registry / Knowledge Base.
B. Integration of Information and Knowledge on FAIR Principles
A re-usable Solution for Knowledge Management C2M2
Knowledge Graph
The ability to access, via a human interface or an API, a vast, semantically-coherent knowledge graph integratedfrom data and relationships generated from different CF projects.
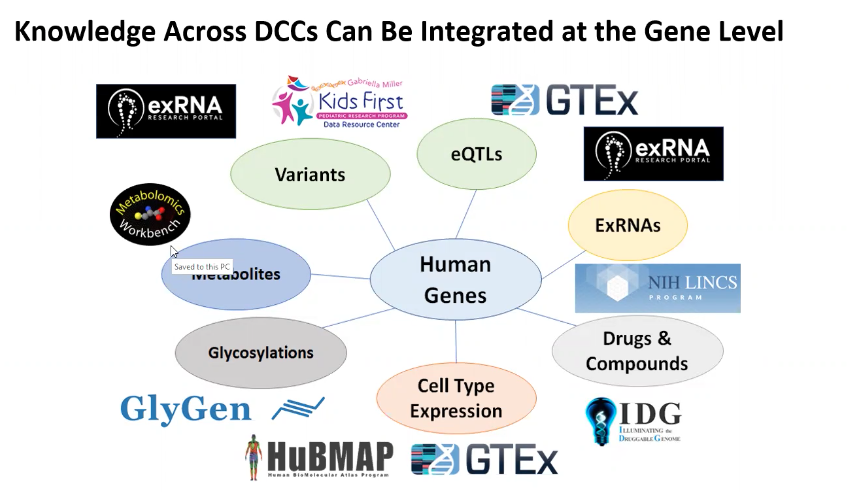
Genes and Variants
A major challenge in studying genes and their associated variants is for scientists to gather such information from scattered data resources. To address this issue, the CFDE Gene-Centric Appyter was developed. An Appyter is a web-based software application that enables researchers to execute bioinformatics workflows without coding. For example, using the Gene-Centric Appyter, researchers enter the gene or variant of interest, and the Appyter queries each CFDE DCCs for information on the gene or variant of interest and returns a summary report.
Drugs and Birth Defects
These videos demonstrate ways for researchers to easily assemble collated and organized information on genes of interest. The Gene and Drug Landing Page Aggregator (GDLPA) pulls together information on genes, drugs, and variants across DCCs. GDLPA provides links to 52 gene-centric landing pages, 18 variant-centric landing pages, and 22 drug-centric landing pages. The CFDE ReproTox Knowledge Graph was developed to harmonize all gene-centric information across CFDE DCCs that produce gene information. A web-based UI allows users to input a gene or gene attribute and generate a network visualization of the relationships between the input entity and available data.
C. Cross-Cutting Discovery Use Cases Catalyzed by Integration across CFDE Projects
Cancer Research
This video describes a precision therapy use case focused on pediatric cancer using RNA-seq data from Kids First and GTEx. Data is imported into the CAVATICA workspace, where users can integrate their data with dbGaP or other data and run various analytical pipelines. The use case notes the challenge of coordinating data across data silos and illustrates how CFDE enabled data harmonization across CFDE DCCs to support knowledge creation and, ultimately, clinical impact.
Assessment of Genetic Variants in Human Disease
FAIR exchange of regulatory information and knowledge This video illustrates how CFDE resources are used to analyze whole genome sequencing (WGS) data for the study of regulatory non-coding variants in congenital heart disease (CHD). Assessing the functional impact of genetic variants remains a significant challenge due to the distributed and inaccessible nature of regulatory variant data, information, and knowledge. Using the approach described, one can examine regulatory non-coding variants associated with CHD by integrating Kids First variants with GTEx xQTL/sQTL data and ERCC regulatory information. In addition, the use case demonstrates the utility of the ClinGen Allele Registry (CAR) by registering and uniquely naming 285M Kids First variants and the Linked Data Hub (LDH) linking and excerpting service to examine the relationship between genes, regulatory elements, and variants.
D. Additional Resources
Overview and Significance of Common Fund Data Ecosystem
Using the Exploration User Interface (EUI) on the GTEx Portal
The GTEx and HuBMAP teams have mapped 16 GTEx tissue locations to the HuBMAP Common Coordinate Framework (CCF). These 16 three-dimensional tissue block locations can be explored via the HuBMAP Exploration User Interface that has been integrated into the GTEx Portal.
Running the Tumor Target Screener Appyter in CAVATICA
The aim of the Tumor Target Screener Appyter is to compare the gene expression patterns in a patient tumor with gene expression patterns in normal tissues and cell types from GTEx transcriptomics and proteomics, ARCHS4, Tabula Sapiens, Human Protein Atlas (HPA), and Human Proteome Map (HPM). The Appyter takes tumor expression data and ranks significantly differentially expressed genes across all tissues and cell types.
Running the xAligner Appyter in CAVATICA
The xAlign Appyter takes as input a fastq file, or a directory containing a collection of fastq files, with raw RNA-seq data. It then loads the files into a cloud environment to run the alignment process on these files to produce gene counts. Several aligners are made available via the Appyter, and the workflow can run in a protected environment such as CAVATICA.
Gene Partnership Knowledge Graph
The Gene Partnership Knowledge Graph (KG) provides access to gene-attribute associations from LINCS (drug-gene), MW (metabolite-gene), IDG (drug-gene), GTEx (tissue-gene), GlyGen (glycan-gene), and BioGRID (gene-gene). The data is stored in a Neo4J database and made accessible via an original knowledge graph interface that we developed specifically for this project. Users can enter into the interface a gene or an attribute to receive a network visualization of the top associations. The interface also supports shortest path search between pairs of entities, as well as various filters for visualization results.
Gene and Drug Landing Page Aggregator
The Gene and Drug Landing Page Aggregator (GDLPA) has links to 53 gene, 18 variant and 19 drug repositories that provide direct links to gene and drug landing pages. Users can search by gene or drug name and then choose the sites that contain knowledge about a gene or drug of interest. Resources supported by the NIH Common Fund are listed first and have the CFDE logo in the top right corner.
Gene-Centric Information across DCC Resources
A majority of Common Fund Data use Gene (or implied Protein) as a key entity to relate to all data in the resource. The objective of developing a gene-centric query across DCCs starting from CFDE CC portal is to provide the research community a user-friendly, intuitive, and interoperable query interface. The Gene query interface provides an ID conversion tool (developed by MW) that seamlessly links all terms used by DCCs and commonly by the user community, an interface on the CFDE-CC page and an appyter that enables the user to link to any DCC that has gene-based data/knowledge, and the DCC landing pages that provide the ability to navigate the data pertinent to the gene(s) within the DCC resources. The gene pages at the portal provide CFDE metadata related information relevant for the gene in question. The gene-partnership appyter facilitates visualization (tables and plots) of DCC-specific information for a gene. The appyter uses APIs provided by DCCs to fetch DCC-specific data relevant to genes. It also provides direct links to DCC-specific landing pages for the gene being searched and is accessible from the gene pages at the CFDE portal. We will illustrate the gene search at the CFDE portal and the gene-partnership appyter through two exemplar genes, PNPLA3 a gene implicated in Caucasian population afflicted with non-alcoholic fatty liver disease, and IDH1 a metabolically associated gene implicated in brain tumor. While these examples show the power of the query system, the user can easily query any gene(s) of interest starting from the CFDE-CC portal.
CFChemDb, CFDE Chemical Database with Chemo-centric Scientific Use Cases
Chemicals are an important entity type for several CF programs and biomedicine in general. Key roles are played by small molecules including drugs, nutrients, toxins, hormones, metabolites, and glycans. Cheminformatics provides rigorous methodology, via powerful and convenient tools, for harmonizing and joining diverse CF and other datasets to explore important scientific use cases, especially in the areas of chemical biology and pharmaceutical discovery. Thus, CFChemDb was developed and comprised of CF chemical datasets and analysis tools to address this need and facilitate discovery via integrative data science combining datasets across the Common Fund Data Ecosystem.
CFDE Resource for Human Subjects Metadata-Driven Data Integration
A major objective of the CFDE is to connect data repositories across DCCs and enable new genomic discoveries through FAIR sharing of data, information, and knowledge. The Variant Partnership has done so by coordinating gene regulatory data and infrastructure across the ExRNA, Kids First, and GTEx DCCs to enable regulatory variant assessment in human diseases. We use the Kids First Data Portal and Variant Workbench to select for disease associated variant data subsets; the ClinGen Allele Registry to register those variants and provide stable and globally unique identifiers for the variants to make them Findable and Accessible; And Linked Data Hub to provide variant associated excerpts from and links to external databases, including GTEx, for regulatory information and knowledge collation. Such regulatory information and knowledge can then be used to inform downstream burden tests for enrichment of likely pathogenic de novo variants in gene regulatory regions and prioritization of likely-disease-causative variants. In combination, the variant partnership bridges the gap between data repositories to enable discoveries of disease-causing regulatory variants.
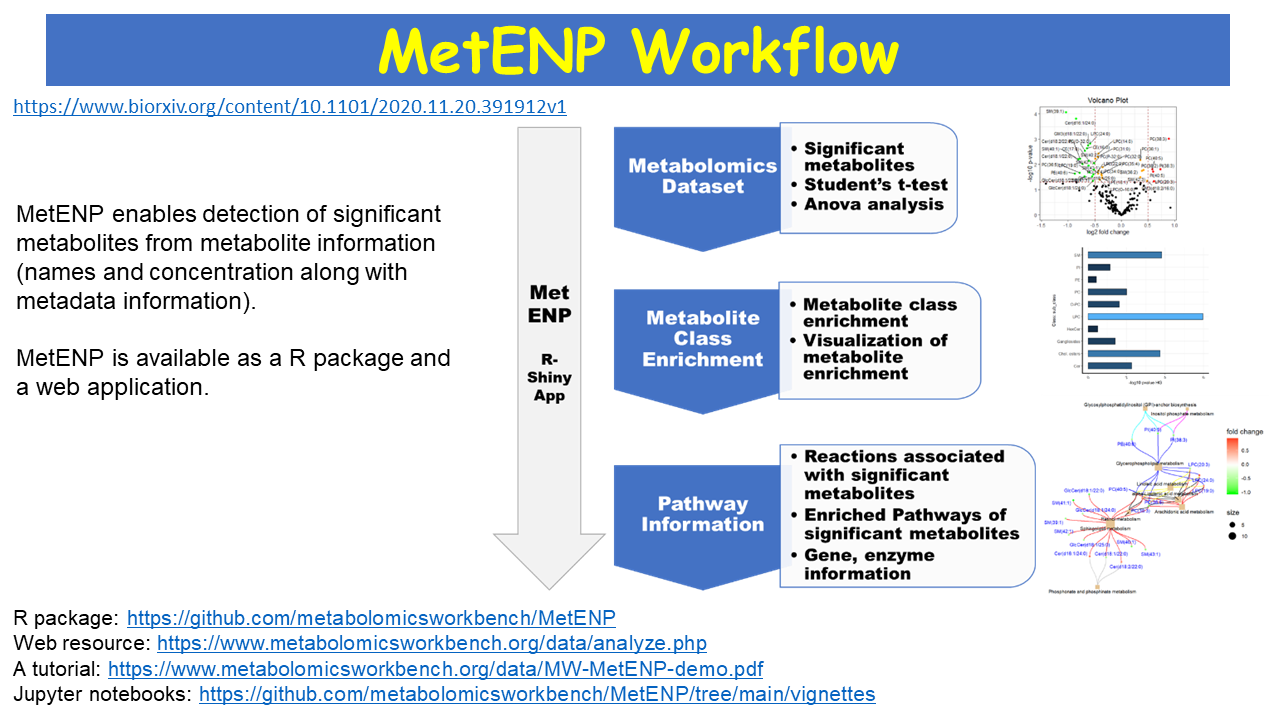
MetENP: A CFDE Workflow Environment for Metabolomics
MetENP is an R package, and a user-friendly web application deployed at the Metabolomics Workbench site that extends the metabolomics enrichment analysis to include species-specific pathway analysis, pathway enrichment scores, gene-enzyme information, and enzymatic activities of the significantly altered metabolites. MetENP provides a highly customizable workflow through various user-specified options and includes support for all metabolite species with available KEGG pathways. MetENPweb is a web application for calculating metabolite and pathway enrichment analysis. MetENP package is freely available from Metabolomics Workbench
E. Appendix
Click here for a complete list of these videos.
To suggest changes, edit this file on GitHub or email the helpdesk.